Overcoming resistance
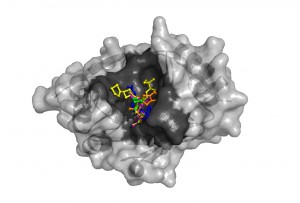
The NDM-1 enzyme’s structure revealed a large cavity (dark gray) capable of binding a variety of known antibiotics (shown in different colors). Once bound, the enzyme can cut the carbapenem ring, destroying the compound’s antibiotic activity. Modeling the interactions computationally can allow researchers to design compounds that will readily adhere to NDM-1 and prevent it from binding with antibiotics. (Argonne National Laboratory.)
As word spread from India last year that common bacteria strains showed resistance to all known antibiotics, some officials feared that terrorists might find a way to weaponize those bacteria and trigger an epidemic. A later study revealed these bacteria were in virtually every modern country’s drinking water supply. Officials worried that if people picked up one of these bugs, became ill and tried taking antibiotics to recover, the bacteria could bypass any drug.
To produce the first publicly available model of a potent enzyme that was making the bacteria into superbugs, groups from across the Department of Energy’s Argonne National Laboratory and collaborators at Texas A&M University plied the computational muscle of Intrepid, the lab’s IBM Blue Gene/P.
Researchers focused on the bacterial enzyme NDM-1 (New Delhi Metallo-β-lactamase), which grabs a piece of the antibiotic beta-lactam ring, breaks it and renders the antibiotic useless. Analysis revealed the enzyme has an enormous and flexible active site, capable of gobbling the stout rings of existing antibiotics, including those of many powerful carbapenems, antibiotics of last resort. Decoding NDM-1 and quickly publishing their findings, the Argonne team documented the enzymes’ promiscuity and uncovered strategic flaws in existing antibiotic development strategies.
“From our simulations it appears that NDM-1 is capable of processing much larger molecules, voiding the easy path” of modifying existing molecules “to breathe new life into old drugs,” says Andrew Binkowski, assistant scientist in Argonne’s biosciences division and a fellow at the University of Chicago’s Computation Institute.
“The NDM-1 story is a really good test case of speed to solution,” says Binkowski, who collaborates with a team from Argonne, the University of Chicago and Northwestern University. “The time to the final solution is really important. If there is an outbreak and we want to quickly find viable drug leads to treat the infection, we can’t afford to wait hundreds of days. We want it in a matter of hours. Intrepid enables that possibility. We went in short order from headlines about a new national security threat to a crystallographic model of the protein used for designing new antibiotics, just months later.”




