Virtual vessels
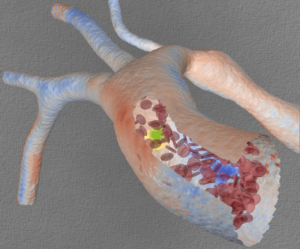
Simulated flow in the human aorta at 20-micron resolution. Blue denotes low wall-shear stress, red denotes high stress. Cutaway: a mockup of red blood cells and a circulating tumor cell (yellow) moving through the vasculature. (Image: Liam Krauss, Lawrence Livermore National Laboratory.)
When a patient shows signs that a major blood vessel wall is weakening, a physician must weigh the risk of surgery against that of the vessel wall’s rupturing catastrophically. That clinical decision is based on experience, images of the vessels and the patient’s other risk factors. But what if a physician could experiment with different surgical options before operating?
Computational modeling of the human circulatory system has arrived, and physicians are eager to test it at Duke University, where Amanda Randles is an assistant professor of biomedical engineering. Randles, a Department of Energy Computational Science Graduate Fellowship (DOE CSGF) alumna, has gained global acclaim for her circulation codes, earning two nominations for the prestigious Gordon Bell Prize for high-performance computing (HPC). Her latest, in 2015, was for HARVEY, a program named for the 16th Century English physician who first described blood circulation. It modeled a fully functioning human arterial system. MIT Technology Review magazine recently named her one of “35 Innovators Under 35” for 2017.
Randles started HARVEY as a Harvard University graduate student. With skills gained while programming IBM’s Blue Gene supercomputer for four years, she modeled patient-specific coronary artery geometries on 294,912 processors.
She says HARVEY might not exist without support from the fellowship, which “allowed me the freedom to pursue a line of research tangential to the main focus of my advisor’s lab.”
Now entering her third year at Duke, Randles has set even more ambitious goals, including modeling the capillary beds that deliver oxygen to tissue and incorporating veins, which return oxygen-depleted blood to the heart. That will likely require more computational power than available on existing HPC systems, so Randles is driving the computer science side of her research.
Several group members are working to port HARVEY to next-generation computing architectures.
These computational challenges serve a larger goal in Randles’ research. As a Duke undergraduate, she majored in both physics and computer science but was drawn to biological science.
“I’ve always wanted to work on medical problems that ultimately could help people,” she says. “Every project we’re working on has a clinician on it, just to make sure we’re asking the right questions.”
Her research uses computational science to evaluate approaches in ways that would be difficult or unethical to try in people. Take the question of whether to operate on a patient developing an aortic aneurysm, a weakening in the central vessel delivering blood from the heart. A physician might make different decisions about whether to line the vessel wall with an artificial support if he or she could visualize areas of weakness and model the potential risk.
Randles wondered if by changing a diseased vessel’s geometry “to represent different treatment options before the doctor ever goes into the operating room, can we see how it would affect the patient under different physiological conditions?”
That scenario is playing out in a project to engage cardiologists in simulated case studies. In Randles’ lab, an interactive model lets physicians see the diseased artery and where HARVEY predicts the vessel wall might undergo low or oscillatory shear stress, leading to dangerous plaque accumulation. The research team wants to know whether that information changes the physician’s decision of where to place a surgical mesh to support the vessel.
To complement research on forces affecting arterial blood flow, Randles is modeling how individual cells twist, fold and deform as they flow through vessels. With support from a National Institutes of Health Early Independence award, she’s using computing to study the transport of individual cancer cells in the bloodstream.
The DOE CSGF, Randles says, made her aware of the Lawrence Fellowship, a program that finances postdoctoral researchers at Lawrence Livermore National Laboratory (LLNL). She won the award in 2013 and began collaborating with LLNL scientists Erik Draeger, Maxim Shusteff, Ted Laurence, Monica Moya and Sonny Ly to model rigid vessel wall geometries compared with three- dimensional physical models.
“We’re moving from the large scale to the small scale,” Randles says.
The team continues collaborating. With support from two laboratory-directed research and development awards, they’re studying fluid flow inside networks of bioprinted tissue – “3-D printed vasculatures made of clear polyurethane to mimic the geometry,” she says. LLNL scientists are assembling sophisticated capillary networks of endothelial cells to provide real-world test beds for validating results Randles obtained running HARVEY on LLNL’s Vulcan HPC system.
Randles is at home working with chips and processors but isn’t afraid – well not too afraid – to dive into the medical side.
“I’ve been in the cardiac catheterization lab,” she says. “I’ve been invited to the operating room, but I’m afraid I might get queasy.”
This article is excerpted from the 2017 DEIXIS: The DOE-CSGF Annual. To see the issue and the rest of the DEIXIS print archive, go here.
About the Author
Karyn Hede is news editor of the Nature Publishing Group journal Genetics in Medicine and a correspondent for the Journal of the National Cancer Institute. Her freelance writing has appeared in Science, New Scientist, Technology Review and elsewhere. She teaches scientific writing at the University of North Carolina, Chapel Hill, where she earned advanced degrees in journalism and biology.




