Targeting tumors
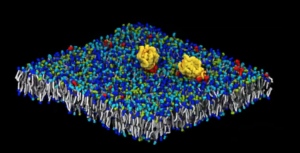
Two RAS proteins (yellow) aggregate atop a complex, eight-lipid-type cell-membrane model in a frame captured from an animation. (Image: Lawrence Livermore National Laboratory.)
Because most cancers are difficult to treat, scientists have long sought the disease’s Achilles’ heel. In some cases, identifying an errant protein or the biological pathway it uses to damage cells could give drug developers a specific target for disrupting tumor growth. But using targeted cancer therapies is often a game of whack-a-mole: Blocking one route with a drug often means that another will compensate to continue cell proliferation and tumor growth.
One notable pathway that has largely evaded therapeutic treatment is the one mediated by the RAS and RAF proteins. RAS and RAF are oncogenes: proteins that can initiate cancer development. Both sit inside human cells and relay signals from outside the cell membrane to the interior. RAS mutations are behind nearly a third of all U.S. cancer diagnoses and are present in nearly all pancreatic cancers, 45 percent of colon cancers and about a third of lung cancers. So far, there has been little success in developing drugs that directly inhibit the RAS pathway.
In 2016, the Department of Energy (DOE) and the National Cancer Institute (NCI) joined to build a large-scale system for a cancer-initiation pilot study that explores RAS-RAF dynamics through supercomputer simulations. Computer scientist Harsh Bhatia at the Center for Applied Scientific Computing at Lawrence Livermore National Laboratory (LLNL) was one of several LLNL researchers to join the effort. Bhatia was up to the challenge even though he hadn’t yet applied computer programming or machine learning to the life sciences.
Together, DOE and NCI developed the Multiscale Machine-Learning Model Infrastructure (MuMMI) to study tumor-initiating pathways. The researchers have demonstrated the technology so far on the two largest U.S. supercomputers: Sierra (LLNL) in 2019 and Summit, at the Oak Ridge Leadership Computing Facility, in 2021. And this month, Bhatia was lead author of a paper on multiscale simulations in Nature Machine Intelligence.
The team has conducted two massive simulation campaigns focusing on RAS and RAF interactions with the cell membrane, using 9.2 million computing hours and generating more than 500 terabytes of data. Bhatia has a 2020 ASCR Leadership Computing Challenge Award to study the RAS-RAF pathway with artificial intelligence methods. ASCR is the DOE’s Advanced Scientific Computing Research program.
Whereas computer scientists typically use machine learning to study how proteins fold and interact, Bhatia and his colleagues study the interaction of multiple proteins. The challenge: to run simulations that can provide high-resolution details of large samples. To enable such studies, MuMMI uses machine learning to couple coarse, large-scale models with precise, small-scale ones. Bhatia says this application of machine learning is novel. “Think of this as an automated computational microscope that analyzes the coarse model and figures out interesting and important regions that warrant a fine-scale simulation. Not only is it a new way of using machine learning but also opens up many new questions with respect to the machine-learning research itself.”
To study molecular pathway with supercomputers, Bhatia and his colleagues first collected existing data from published papers on how different RAS-RAF pathway components initiate cancer. Running this simulation generated different configurations of what proteins interact and how they’re arranged. MuMMI also simultaneously generated molecular and atomic-scale details using a new machine-learning based framework.
Machine learning, Bhatia explains, groups protein interactions by similarity or difference. “If you run it long enough, you can see an event that occurs once in a millisecond,” he says. “That’s still quite frequent for the human body, and you don’t want to miss it. But a millisecond already approaches the limits of what can be studied computationally.” Ultimately, machine learning helps simulate all the protein interaction possibilities.
After the first set of simulations, Bhatia and his colleagues have zoomed in on how the RAS protein re-arranges lipids, major cell membrane components. Now they’re also studying RAF, the next protein in the RAS pathway. The researchers used MuMMI to study how RAS and RAF interact with one another and with the membrane. “We’re asking the same questions, but at a grander scale, with more complexity,” Bhatia says. “All these simulations are not independent; they are all connected by machine learning and the more accurate simulations are used to improve the quality of the coarser ones.”
Bhatia and his colleagues work with the NCI-operated Frederick National Laboratory for Cancer Research to validate the computational output. One of the most notable findings from early results captured the wide range of RAS protein interactions from tens of thousands of unique simulations, Bhatia says.
The researchers will examine different proteins in the RAS family and mutations that may affect protein and lipid dynamics, seeking hints that cancer biologists and drug developers might use to design better therapies – which will, in turn, lead to further advances in computing.
“Without a really important problem, we would not have been able to make this progress from the computational side,” Bhatia says. “And without these kinds of computational capabilities, we wouldn’t be able to make progress on the (biology) side. That synergy is what’s really exciting about the project.”
About the Author
Wudan Yan is a Seattle-based freelance science writer.




